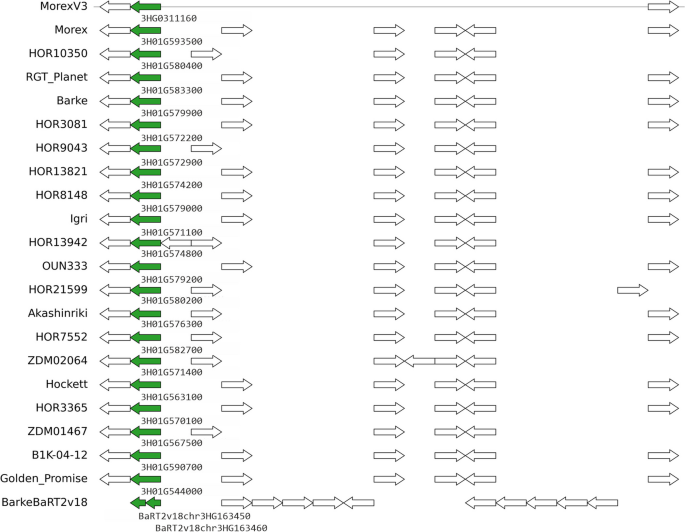
barley_pangenes
Pangenes are clusters of gene models/alleles found in barley assemblies in a similar genomic location.
Click on chromosomes below to browse pangenes and display multiple sequence alignments (MSA) of encoded proteins. Read our blog post for more information.
| chr1H | chr2H | chr3H | chr4H | chr5H | chr6H | chr7H | chrUn |
Also, MorexV3 HC genes in maps produced by BARLEYMAP link out to these MSAs.
Raw files and methods
You can download the raw pangene matrices and get more details at https://github.com/eead-csic-compbio/barley_pangenes.
Citations
Protocol first published at:
Contreras-Moreira B, Saraf S, Naamati G et al (2023) GET_PANGENES: calling pangenes from plant genome alignments confirms presence-absence variation. Genome Biol 24, 223. https://doi.org/10.1186/s13059-023-03071-z
Multiple sequence alignments produced with clustal-omega v1.2.4:
Sievers F, Wilm A, Dineen D, et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038%2Fmsb.2011.75
Annotated genomes from:
Jayakodi, M., Padmarasu, S., Haberer, G. et al. The barley pan-genome reveals the hidden legacy of mutation breeding. Nature 588, 284-289 (2020). https://doi.org/10.1038/s41586-020-2947-8
Mascher M, Wicker T, Jenkins J, et al. Long-read sequence assembly: a technical evaluation in barley, The Plant Cell, Volume 33, Issue 6, June 2021, Pages 1888-1906, https://doi.org/10.1093/plcell/koab077
Coulter M, Entizne JC, Guo W, et al. BaRTv2: a highly resolved barley reference transcriptome for accurate transcript-specific RNA-seq quantification. Plant J. 2022 111(4):1183-1202. https://doi.org/10.1111%2Ftpj.15871
Funding
[PRIMA GENDIBAR, PCI2019-103526] supported by Horizon 2020
[A08_23R] funded by Aragón goverment
[FAS2022_052, INFRA24018, conexión BCB] funded by CSIC
[PID2022-142116OB-I00] by MICIU/AEI/10.13039/501100011033
