coexpression_motif_discovery
This repository documents recipes to discover cis-regulatory motifs within proximal promoters of plants in RSAT::Plants.
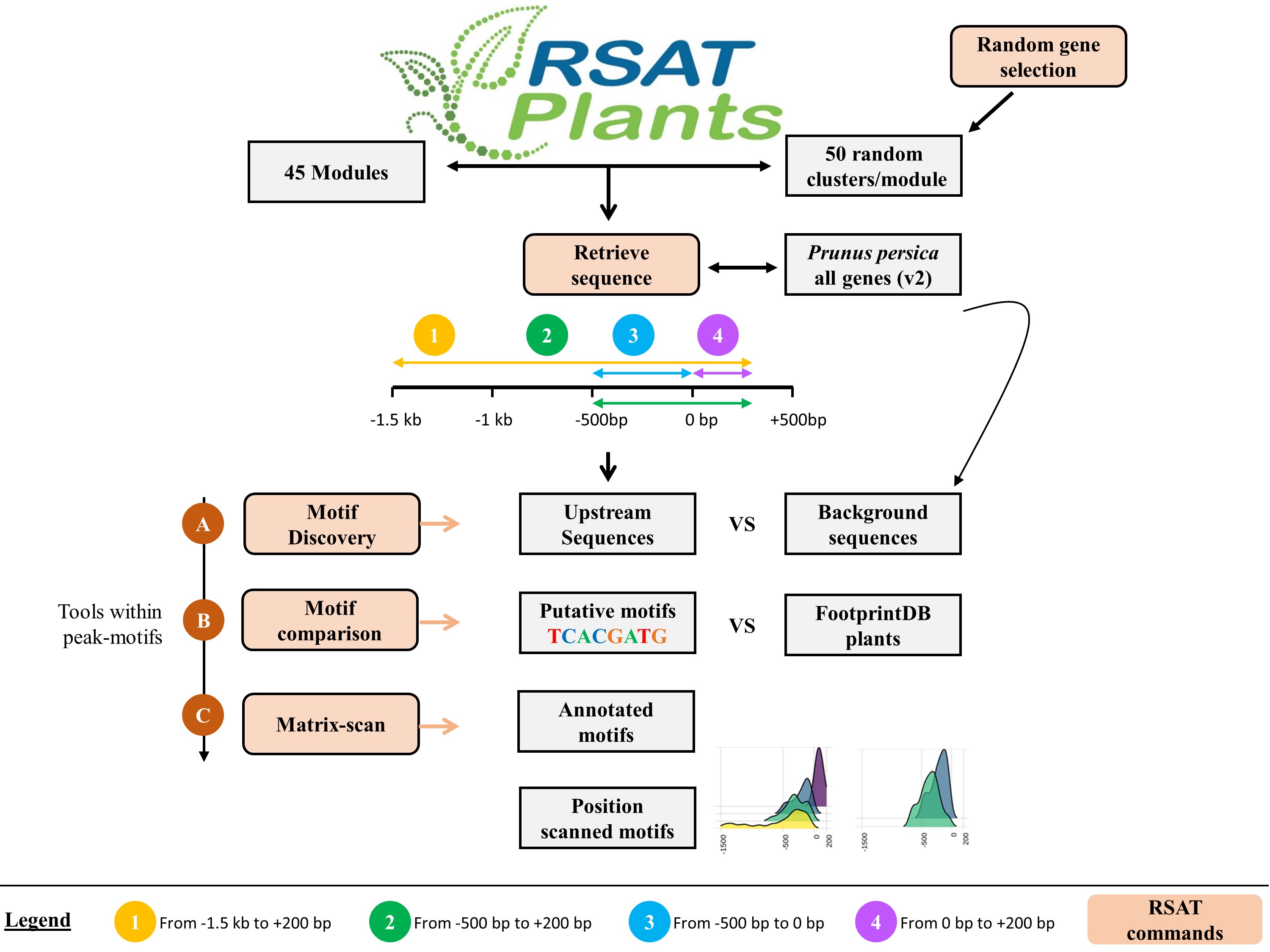
The reports and data in folder peach/ correspond to the benchmark experiment with peach expression-based gene modules. You can browse them at eead-csic-compbio.github.io/coexpression_motif_discovery/peach .
The recipes can be executed on the command-line with on a Docker container, as explained in the tutorial, or using the RSAT Web interfaces, as explained in the Web protocol.
Citation: N Ksouri(1), JA Castro-Mondragón (2,3), F Montardit-Tardà (1), J van Helden (2), B Contreras-Moreira (1,4,5), Y Gogorcena (1) (2021) Tuning promoter boundaries improves regulatory motif discovery in non-model plants: the peach example. Plant Physiology, https://doi.org/10.1093/plphys/kiaa091
Affiliations
- Estación Experimental de Aula Dei-CSIC, Zaragoza, Spain
- Aix-Marseille Univ, Theory and Approaches of Genome Complexity (TAGC), Marseille, France.
- Centre for Molecular Medicine Norway (NCMM), Nordic EMBL Partnership, University of Oslo, Norway.
- Fundacion ARAID, Zaragoza, Spain
- European Bioinformatics Institute EMBL-EBI, Hinxton, UK
Questions or comments, please contact: nksouri at eead.csic.es
Docker Biocontainer
A docker biocontainer with Regulatory Sequence Analysis Tools (RSAT) image is available here. If you have not set a docker engine on your machine, please see the instructions provided by the docker community for a simplified installation procedure.
Once docker is set up, you can get and run the RSAT docker image by typing the following command lines in the terminal:
# 1. Get the docker image (This will take 10GB of your filesystem)
# from the Linux/WSL terminal
$docker pull biocontainers/rsat:20240507_cv1
# 2. Create local folders for input data and results named respectively rsat_data/ and rsat_results/
# In addition, a subfolder (rsat_data/genomes) should be created too
$mkdir -p $HOME/rsat_data/genomes rsat_results
$chmod -R a+w rsat_data rsat_results
# 3. Launch Docker RSAT container
$docker run --rm -v $HOME/rsat_data:/packages/rsat/public_html/data/ -v $HOME/rsat_results:/home/rsat_user/rsat_results -it biocontainers/rsat:20240507_cv1
# you should see a warning that can be safely ignored:
# * Starting Apache httpd web server apache2
# (13)Permission denied: AH00091: apache2: could not open error log file /var/log/apache2/error.log.
# AH00015: Unable to open logs
# Action 'start' failed.
# The Apache error log may have more information
# 4. Download organism from public RSAT server (in this case Prunus persica)
$download-organism -v 2 -org Prunus_persica.Prunus_persica_NCBIv2.38 -server https://rsat.eead.csic.es/plants
# 5. Test the container
$cd rsat_results
$make -f ../test_data/peak-motifs.mk RNDSAMPLES=2 all
“rsat_results” and “rsat_data” are two local directories in the host machine serving as a persistant storage volume inside the RSAT docker container.
- The dockerfile used to create this Biocontainer is available here
Funding
This work was partly funded by the Spanish Ministry of Economy and Competitiveness grants AGL2014-52063R, AGL2017-83358-R (MCIU/AEI/FEDER/UE); and the Government of Aragón with grants A44 and A09_17R, which were co-financed with FEDER funds. N Ksouri was hired by project AGL2014-52063R and now funded by a PhD fellowship awarded by the Government of Aragón and co-financed with ESF funds respectively.
